NCBI stands for the National center for biotechnology information which is a composite database. We already have discussed the databases (Read here). The URL for NCBI is https://ncbi.nlm.nih.gov
NCBI is an important database for biotechnology students and researchers. There are various tools available for the analysis of data. NCBI is inside connected with other databases such as GenBank, PubMed.
NCBI is a part of the united states national library of medicine (NLM) and the National Institute of health. NCBI is in Bethesda, Maryland founded in 1988 by senator Claude pepper.
NCBI along with EBI and CIB is a member of INSDC (international sequence database collaboration). The three members of INSDC are DDBJ (DNA databank of japan), NCBI (national center for biotechnology information), EMBL (European molecular biology laboratory). Every member of INSDC shares its information every 24 hr. so we can access similar data from any of these databases.
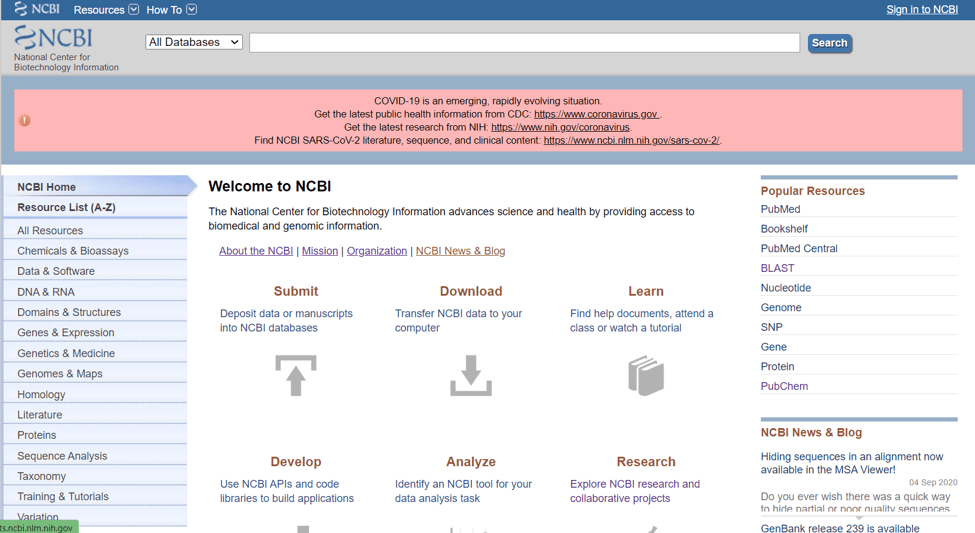
(Fig.1 NCBI homepage)
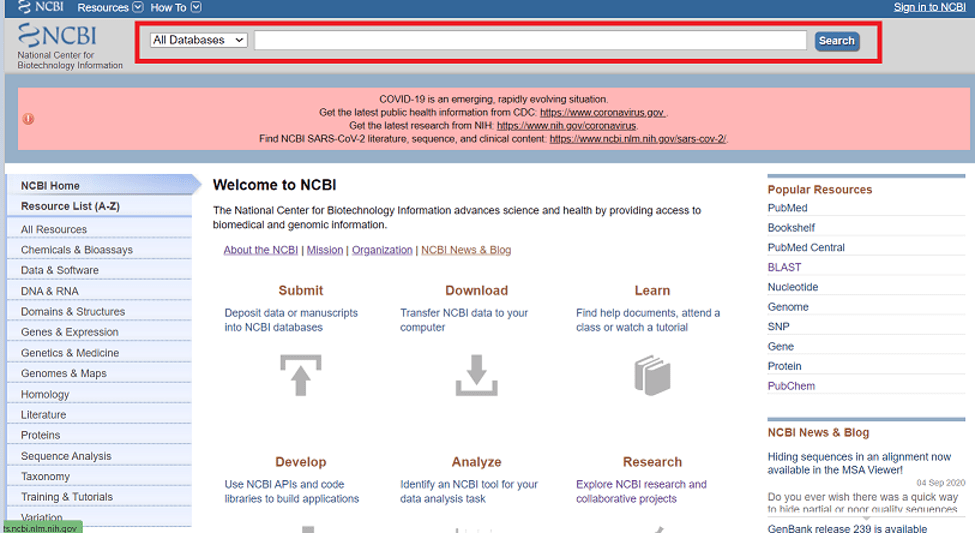
NCBI has its search engine known as Entrez (Fig.2 search engine of NCBI)
Entrez is an integrated data retrieval system, which provides access to a diverse set of more than 40 databases contains 690 million records. Entrez allows users to search using text or Boolean queries, to download the data in various formats.
We know that NCBI is a composite database that we can see it in the above picture.
How to retrieve data from NCBI?
Data retrieval from NCBI is an easy process.
We are going the search for keratin protein sequence in the NCBI and get the fasta from there.
- Step 1: Go to NCBI homepage
- Step 2: Search for the keratin in the Entrez. Here you can also select which database you want to search for your query for example keratin is a protein so you can select the protein database, or you can leave it to search within all databases.
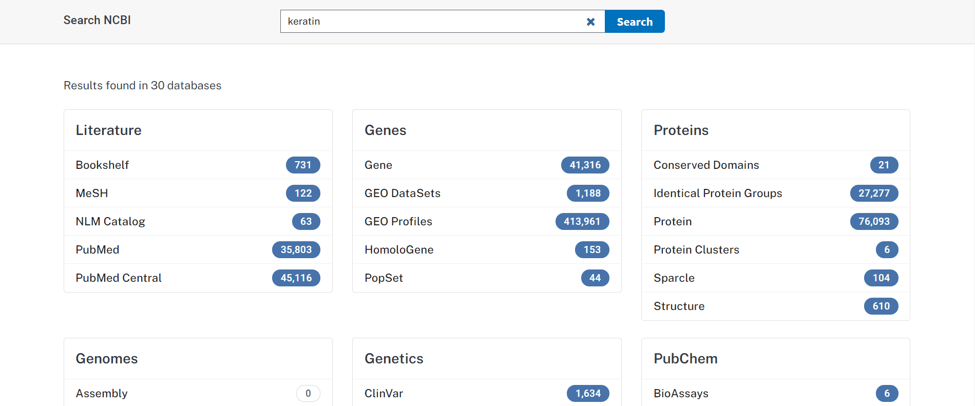
- Step 3: Now select the protein
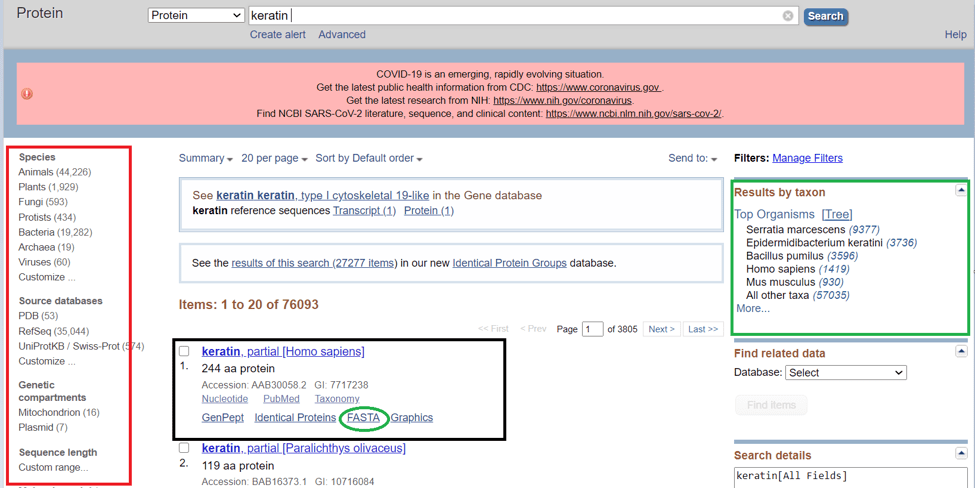
- Step 4: if you are looking for the fasta of keratin you can just select it from the searched results. Or you can differentiate your results with provided options. For example, you can differentiate your result by taxon (on the right corner within the green box in the above picture.)

How to search for a sequence that is unknown to us?
If you have a protein or nucleotide sequence that you want to look for in the database. You can use the built-in BLAST tool.
- Go to the NCBI homepage and look for BLAST on the right corner of the Homepage (Fig1).
- Then select the nucleotide or protein BLAST.
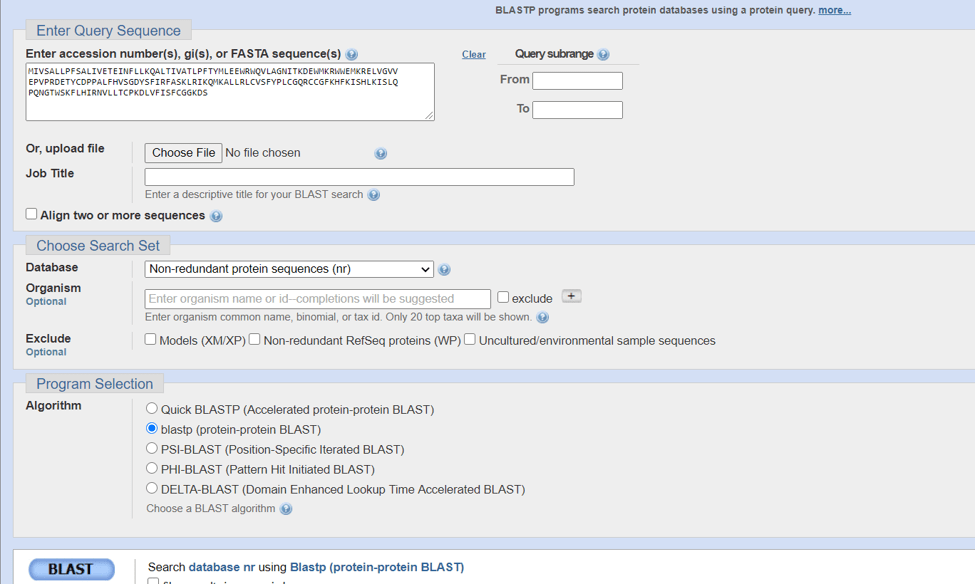
- Put the sequence in the query box and hit BLAST on the bottom. Here you can also select which organism data you want to search. And you can give your query a name it is quite helpful when you are working with various sequences.
- I have separated the search result in two pictures.
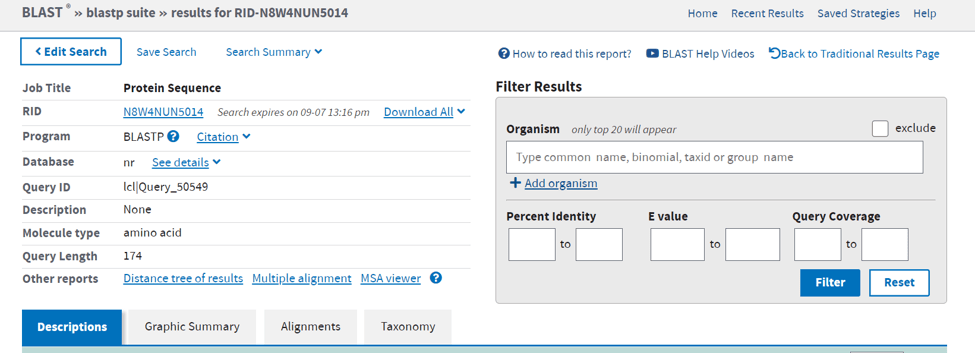
- in this picture, you can see basic information about your queries such as the length of the sequence or you can filter your results.
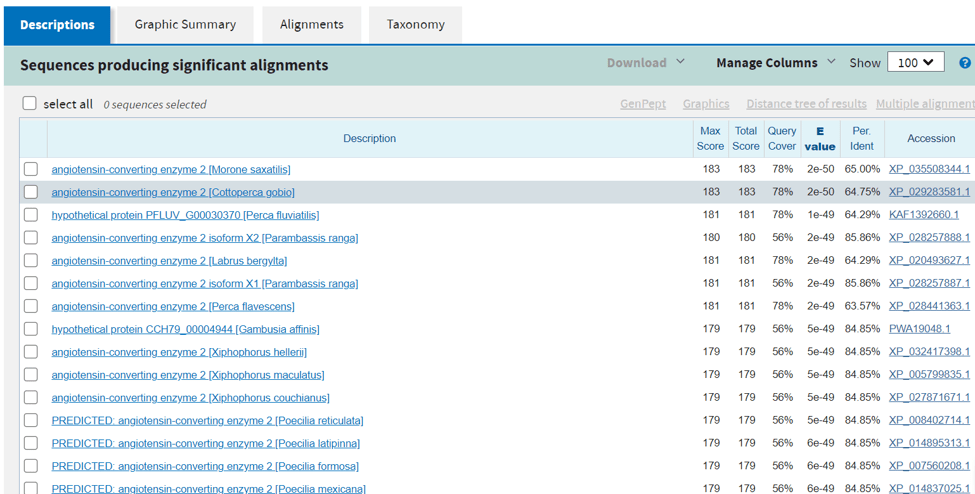
- in this picture, we can see the result of our query and from here we can also check how much percent our sequence is matching to the searched results.
References
- NCBI Resource Coordinators. (2015). Database resources of the national center for biotechnology information. Nucleic acids research, 43(D1), D6-D17.
- Madden, T. (2013). The BLAST sequence analysis tool. In The NCBI Handbook [Internet]. 2nd edition. National Center for Biotechnology Information (US).
- Baxevanis, A. D. (2006). Searching the NCBI databases using Entrez. Current protocols in human genetics, 51(1), 6-10.
When someone writes an post he/she keeps the idea of
a user in his/her mind that how a user can understand it.
Thus that’s why this piece of writing is amazing.
Thanks!
I’d like to thank you for the efforts you have put in penning this site.
I’m hoping to check out the same high-grade content by you in the future as well.
In fact, your creative writing abilities has inspired me to get
my very own blog now 😉
There’s definately a great deal to find out about this topic.
I like all of the points you’ve made.